MNE Python
MNE-Python is a Python package for analysing electrophysiology (MEG, EEG, sEEG, ECoG, NIRS, etc) data.
MNE-Python Versions
Bear Apps has several versions of MNE-Python as modules.
Bear Modules
The following bash loads mne version 1.3.1 and its dependencies - an equivalent is availiable for JupyterLab.
Note that MNE-Python depends on a number of other python applications that will be loaded automatically. The above code will also load numpy, scipy, numba, matplotlib, sklearn and many other packages that are needed by MNE into your environment.
MNE in a virtual environment
If you want to use a specific version of MNE-Python that isn't supported by BEAR, you can install it into a virtual environment. This bash script provides an example. We load Python 3.9.5, create an environment and install MNE into the environment using pip.
#!/bin/bash
module purge;
module load bear-apps/2022a
module load MNE-Python/1.3.1-foss-2022a
module load IPython/7.25.0-GCCcore-10.3.0
export VENV_DIR="${HOME}/virtual-environments"
export VENV_PATH="${VENV_DIR}/mne-example-${BB_CPU}"
# Create master dir if necessary
mkdir -p ${VENV_DIR}
echo ${VENV_PATH}
# Check if virtual environment exists and create it if not
if [[ ! -d ${VENV_PATH} ]]; then
python3 -m venv --system-site-packages ${VENV_PATH}
fi
# Activate virtual environment
source ${VENV_PATH}/bin/activate
# Any additional installations
pip install mne==1.1.0
As with other examples, this can be copied directly into the terminal or saved as an shell script that can be executed in a terminal.
Evoked response example
The following code is adapted from the MNE-Python overvew of MEG/EEG analysis tutorial. It will download a small example file and run a quick analysis.
You can run this code directly in a Python session on BEAR within an activated MNE environment. The file will be saved into your RDS home directory (eg /rds/homes/q/quinna) unless specified otherwise - please change the sample_data_folder variable if you'd like to save this file elsewhere.
import numpy as np
import mne
# This will download example data into your RDS home directory by default -
# change the next line if you want to save the file elsewhere!
sample_data_folder = '/rds/homes/q/quinna/mne-data/'
sample_data_raw_file = (sample_data_folder / 'MEG' / 'sample' /
'sample_audvis_filt-0-40_raw.fif')
raw = mne.io.read_raw_fif(sample_data_raw_file)
events = mne.find_events(raw, stim_channel='STI 014')
event_dict = {'auditory/left': 1, 'auditory/right': 2, 'visual/left': 3,
'visual/right': 4, 'smiley': 5, 'buttonpress': 32}
reject_criteria = dict(mag=4000e-15, # 4000 fT
grad=4000e-13, # 4000 fT/cm
eeg=150e-6, # 150 µV
eog=250e-6) # 250 µV
epochs = mne.Epochs(raw, events, event_id=event_dict, tmin=-0.2, tmax=0.5,
reject=reject_criteria, preload=True)
conds_we_care_about = ['auditory/left', 'auditory/right',
'visual/left', 'visual/right']
epochs.equalize_event_counts(conds_we_care_about) # this operates in-place
aud_epochs = epochs['auditory']
vis_epochs = epochs['visual']
vis_evoked = vis_epochs.average()
fig = vis_evoked.plot_joint()
fig[0].savefig('my-mne-evoked-example-grad.png')
fig[1].savefig('my-mne-evoked-example-mag.png')
fig[2].savefig('my-mne-evoked-example-eeg.png')
We can save this as 'mne_python_example.py` as our core analysis script. This can be execute from a terminal session in which the appropriate python environment has been loaded. For example, we could open a 'BlueBEAR GUI' session, open a new terminal, change directory to the location of our script and run the following code:
At the end you should have some new figures created next to your script.
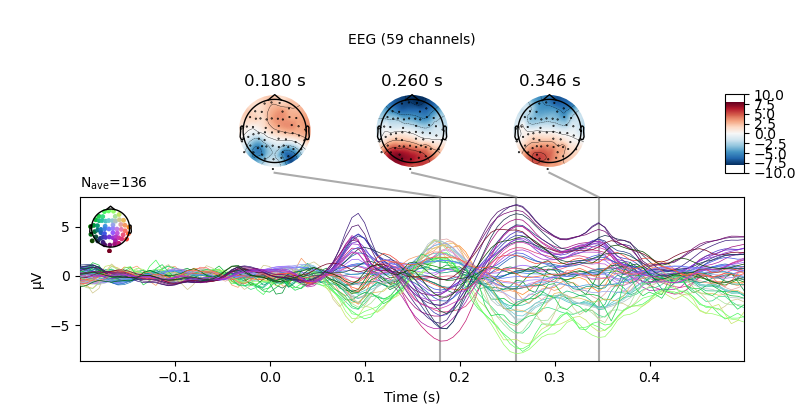
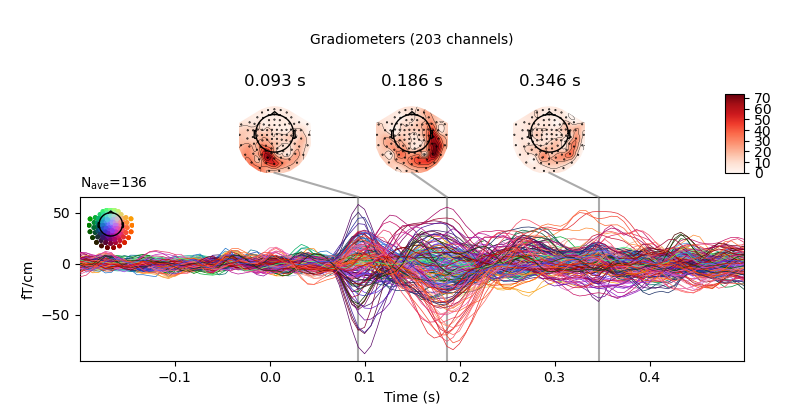
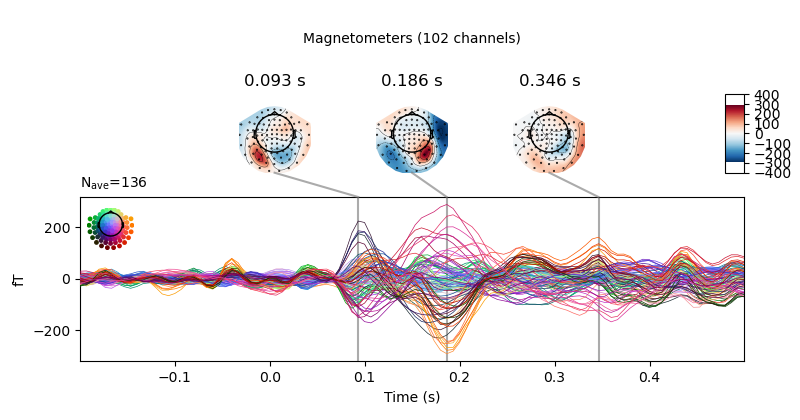
MNE on the cluster example
Warning
There is currently a bug with this example to do with the automatic file downloading when running on the cluster. Running a simliar example on your own data fromo RDS should work fine.
Alternatively, we can run our evoked responses analysis on the cluster. For this we'll need a job submission script. Let's use the following shell code to load the MNE module from BEAR and run our code.
#!/bin/bash
#SBATCH --account quinna-example-project
#SBATCH --qos bbdefault
module purge; module load bluebear
module load bear-apps/2022a
module load MNE-Python/1.3.1-foss-2022a
python mne_python_example.py
Note
We could equally create or load a virtual environment with a customised Python set-up in our job script. Here we use the built in BEAR module as it is all we need for the case in hand.
We can save the job script as run_mne_python_example.sh and submit it to the cluster using sbatch
You can see the progress of the job using the 'Active Jobs' page on BEAR portal or by reading the log files.